Laboratory of Molecular Biology
PROFILE
Our research is focused on the analysis of molecular mechanisms responsible for DNA replication in bacterial cells. We mainly investigate the mechanisms responsible for the initiation and regulation of DNA replication of bacterial plasmids an antibiotic resistance factors in bacterial cells. Model used in our investigations is RK2 plasmid that belongs to the group of broad-host-range replicons that can be stably maintained and transferred between cells of various bacterial species, including plant, animal and human pathogens. The research utilizing such a unique system, allowing for the analysis of cellular mechanisms in various organisms, makes the significant contribution to-wards the general understanding of the fundamental cellular processes im-portant for future development of new antimicrobial therapeutic strategies.
In our investigations we focus on (i) multi-molecule complexes formed on DNA by replication initiation proteins (Rep), their structural bases, functions and dynamics, (ii) analysis of the mechanism of protease activity on substrates bound to DNA and effects of proteases on DNA replication regulation and stably maintenance of plasmids, and (iii) analysis of the function of plasmid encoded post segregational killing systems in the context of its components’ interactions with DNA and their stability.
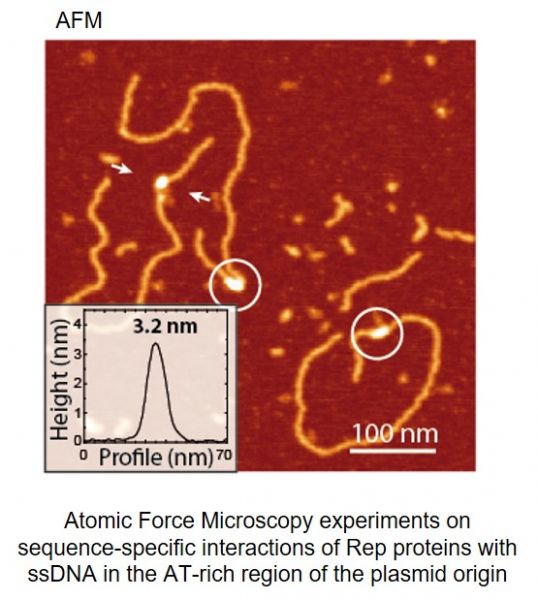
Current understanding of structure and functions of multi-protein complex-es formed on DNA is still limited. Especially challenging is the analysis of in-trinsically disordered proteins and their complexes with single-stranded DNA (ssDNA). After discovery that Rep proteins bound both double-stranded DNA (dsDNA) and similar, as chromosomal DnaA, they also interact with ssDNA (Wegrzyn K. et al., Nucleic Acids Res. 2014), we try to structurally character-ize this complex. In our studies we are applying in vivo and in vitro experi-ments including reconstituted DNA replication assays, real time kinetics of protein-DNA complexes, Mass Spectrometry (MS), Atomic Force Microscopy (AFM) and crystallographic analysis.
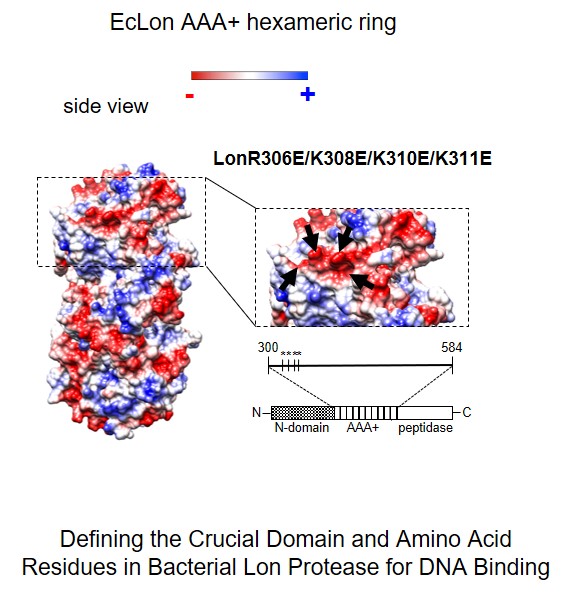
In our research we also challenge the question about the molecular mecha-nism(s) of proteases activity modulation by interaction with nucleic acids. The controlled proteolysis is a long term task as a potential tool in devel-oping anti-microbial therapies. Recently we extended our investigations on the role of proteases in nucleoprotein complex dynamics in bacterial cells. By constructing mutants of Escherichia coli Lon protease, using bioinformatics and phenotypic analysis we demonstrated engagement of positively charged amino acid residues, located on the ATPase domain surface, in interaction with DNA. In vivo tests revealed essentiality of Lon interaction with DNA for Lon activity associated with cell division (Karłowicz A., Węgrzyn K. et. al., JBC 2017).
In the same lane of investigations we tested the influence of proteases on handcuff complex responsible for controlling DNA replication and stable maintenance of plasmids in bacterial cells. We found that bacterial proteas-es, particularly Lon and ClpAP, disrupt handcuff by degrading Rep proteins interacting DNA, thus affecting plasmid stability. Moreover, we demonstrate that Rep monomers are able to dissociate handcuffed plasmid molecules and re-initiate DNA synthesis (Bur K. et. al., NAR 2017). We assume that those mechanisms are important for plasmid maintenance.
We are also developing the line of research considering the mechanisms of replication complex formation in bacterial cells. Our study on the replisome assembly at plasmid RK2 replication origin (Wawrzycka A, et al., Proc Natl Acad Sci USA. 2015) revealed that specific interaction between the plasmid Rep protein and β subunit of DNA Polymerase III holoenzyme (β-clamp) is re-quired for polymerase complex assembly at plasmid origin. Currently conduct-ed experiments are directed towards understanding of the regulatory mecha-nisms involving β-clamp interaction with host and plasmid encoded proteins. We are directing our investigations towards the analysis of the stability of an essential replication proteins and mechanisms involving protease activities for the regulation of both chromosomal and plasmid DNA replication in stress conditions.
Recent and Important Awards:
| 2016 | ELSEVIER-Plasmid prize award for dr. Katarzyna Węgrzyn for the best poster presentation during Plasmid Biology Conference, Cambridge |
| 2015 | Award from The Committee of the Cell Molecular Biology of Polish Academy of Science (PAN) for the best publication in micro-biology published in Polish laboratories |
| 2002 | HHMI (Howard Hughes Medical Institute) Young Investigator |
| 2000 | EMBO YIP (Young Investigator Programme), EMBO Heidelberg |
ONGOING RESEARCH PROJECTS
SONATA NCN (194 172 EUR) started in February 2018
Structural and functional analysis of nucleoprotein complexes of Rep plasmid proteins and the ssDNA of the DUE origin region
The initiation of DNA replication of extrachromosomal genetic elements, plasmids, is induced by Rep proteins. These proteins contain WH (Winged Helix) domains, which are responsible for dsDNA binding. Our research showed that Reps bind specific region of ssDNA of DUE. However, the structure of the nucleoprotein complexes of Reps and the ssDNA DUE is unknown. The aim of the project is the structural-functional analysis of complexes of plasmid replication initiation proteins, Rep, and the ssDNA DUE. Within this project we plan to determine the domains and the amino acid residues of Rep proteins which are responsible for binding of the ssDNA DUE and to investigate the role of ssDNA DUE binding by Reps. The obtained results should allow to acquire structural data on the formation of nucleoprotein complexes by plasmid replication initiators and to broaden our knowledge concerning the process of DNA replication. Therefore, they will have an impact on the development of the field and might also, in further perspective, have great impact on the development of new antimicrobial strategies.
TEAM FNP (808 874 EUR) started in October 2018
Condition-dependent protease activation for targeted proteolysis in the regulation of DNA replication
The presented project is directed at elucidation how, depending on conditions, proteases are structurally and spatially regulated to degrade essential replication proteins. The research will concentrate on bacterial Lon protease and its phosphate containing cofactors, contributing to the protease activities. We will analyze Lon from bacterial species not being investigated so far in this aspect as well as known and newly identified Lon substrates essential for replication of bacterial DNA. By applying in vitro and in vivo experiments, including real-time, single-molecule and single-cell analysis with a variety of Lon proteins, substrates and cofactors, we expect to describe species-specific, substrate-specific and general effects on Lon, and relate them to growth conditions. The cutting-edge AFM and cryo-EM will be utilize for the description of Lon complexes with cofactors and substrates. Our investigations can provide data on Lon as a new target for new antimicrobial strategies.
RESEARCH TEAM
Laboratory of Molecular Biology
Department of Molecular and Cellular Biology
Intercollegiate Faculty of Biotechnology UG&MUG
Abrahama 58 Street, 80-307 Gdańsk, Poland
Office - Stanisława Urbańska
Tel.:+48 58 523 6421
STAFF LIST
Head: prof. dr hab. Igor Konieczny [link]
tel. +48 58 523 6365 [e-mail]
Post doctoral fellows:
dr n. med. inż. Dorota Pomorska [link]
+48 58 523 6351 [e-mail]
dr Katarzyna Węgrzyn [link]
tel. +48 58 523 6325 [e-mail]
dr Tomasz Chamera [link]
tel. +48 58 523 6325 [e-mail]
dr Katarzyna Bury [link]
tel. +48 58 523 6351 [e-mail]
PhD students:
MSc Monika Oliwa (Ciuksza) [e-mail]
MSc Magdalena Sroka [e-mail]
MSc Ewelina Wysocka [e-mail]
tel. +48 58 523 6325
SELECTED PUBLICATIONS
-
Bojko Magdalena, Węgrzyn Katarzyna, Sikorska Emilia, Kocikowski Mikołaj, Parys Maciej, Battin Claire, Steinberger Peter, Kogut Małgorzata, Winnicki Michał, Sieradzan Adam, Spodzieja Marta, Rodziewicz-Motowidło Sylwia. Design, synthesis and biological evaluation of PD-1 derived peptides as inhibitors of PD-1/PD-L1 complex formation for cancer therapy. Bioorganic Chemistry 2022, 128: 106047 (doi: 10.1016/j.bioorg.2022.106047)
-
Kadziński Leszek, Łyżeń Robert, Bury Katarzyna, Banecki Bogdan. Modeling and optimization of β-galactosidase entrapping in polydimethylsiloxane-modified silica composites. International Journal of Molecular Sciences 2022, 23(10): 5395 (doi: 10.3390/ijms23105395)
-
Kuncewicz Katarzyna, Battin Claire, Węgrzyn Katarzyna, Sieradzan Adam, Wardowska Anna, Sikorska Emilia, Giedrojć Irma, Smardz Pamela, Pikuła Michał, Steinberger Peter, Rodziewicz-Motowidło Sylwia, Spodzieja Marta. Targeting the HVEM protein using a fragment of glycoprotein D to inhibit formation of the BTLA/HVEM complex. Bioorganic Chemistry 2022, 122: 105748 (doi: 10.1016/j.bioorg.2022.105748)
- Węgrzyn Katarzyna, Konieczny Igor. Archaeal Orc1 protein interacts with T-rich single-stranded DNA. BMC Research Notes 2021, 14(1): 275 (doi: 10.1186/s13104-021-05690-w)
-
Kołodziej-Sobocińska Marta, Trojanowski Damian, Bury Katarzyna, Hołówka Joanna, Matysik Weronika, Kąkolewska Hanna, Feddersen Helge, Giacomelli Giacomo, Konieczny Igor, Bramkamp Marc, Zakrzewska-Czerwińska Jolanta. Lsr2, a nucleoidassociated protein influencing mycobacterial cell cycle. Scientific Reports 2021, 11(1): 1-12 (doi: 10.1038/s41598-021-82295-0)
-
Kołodziej-Sobocińska Marta, Łebkowski Tomasz, Płociński Przemysław, Hołówka Joanna, Paściak Mariola, Wojtaś Bartosz, Bury Katarzyna, Konieczny Igor, Dziadek Jarosław, Zakrzewska-Czerwińska Jolanta. Lsr2 and its novel paralogue mediate the adjustment of Mycobacterium smegmatis to unfavorable environmental conditions. mSphere 2021, 6(3): 1-18 (doi: 10.1128/msphere.00290-21)
-
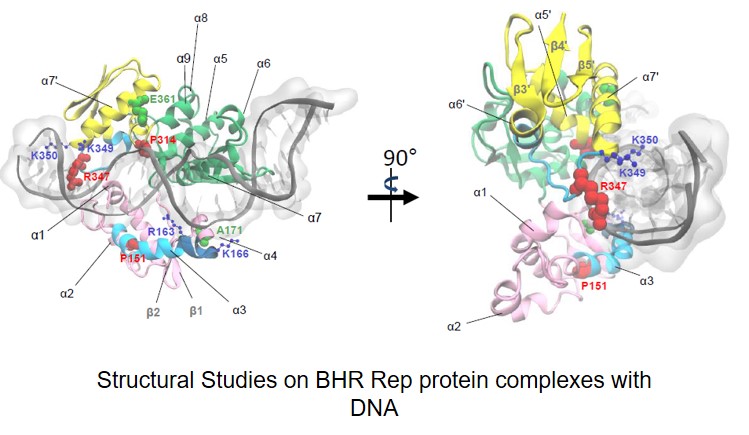
Węgrzyn Katarzyna, Zabrocka Elżbieta, Bury Katarzyna, Tomiczek Bartłomiej, Wieczór Miłosz, Czub Jacek, Uciechowska-Kaczmarzyk Urszula, Moreno-del Alamo Maria, Walkow Urszula, Grochowina Igor, Dutkiewicz Rafał, Bujnicki Janusz M., Giraldo Rafael, Konieczny Igor. Defining a novel domain that provides an essential contribution to site-specific interaction of Rep protein with DNA. Nucleic Acids Research 2021, 49(6): 3394-3408 (doi: 10.1093/nar/gkab113)
-
Romanowska Anita, Węgrzyn Katarzyna, Bury Katarzyna, Sikorska Emilia, Gnatek Aleksandra, Piwkowska Agnieszka, Konieczny Igor, Lesner Adam, Wysocka Magdalena. Novel cell permeable polymers of N-substituted L-2,3-diaminopropionic acid (DAPEGs) and cellular consequences of their interactions with nucleic acids. International Journal Of Molecular Sciences 2021, 22(5): 2571 (doi: 10.3390/ijms22052571)
-
Ropelewska Małgorzata, Gross Marta H., Konieczny Igor. DNA and Polyphosphate in Directed Proteolysis for DNA Replication Control. Frontiers in Microbiology 2020, 11: 585717 (doi: 10.3389/fmicb.2020.585717)
-
Gross H. Marta, Konieczny Igor. Polyphosphate induces the proteolysis of ADP-bound fraction of initiator to inhibit DNA replication initiation upon stress in Escherichia coli. Nucleic Acids Research 2020, 48(10): 5457-5466 (doi: 10.1093/nar/gkaa217)
-
Mróz Dagmara, Wyszkowski Hubert, Szablewski Tomasz, Zawieracz Katarzyna, Dutkiewicz Rafał, Bury Katarzyna, Wortmann Saskia B., Wevers Ron A., Ziętkiewicz Szymon. CLPB (caseinolytic peptidase B homolog), the first mitochondrial protein refoldase associated with human disease. Biochimica et Biophysica Acta (BBA) - General Subjects 2020, 1864 (4): 129512 (doi: 10.1016/j.bbagen.2020.129512)
-
Spodzieja Marta, Kuncewicz Katarzyna, Sieradzan Adam, Karczyńska Agnieszka, Iwaszkiewicz Justyna, Cesson Valérie, Węgrzyn Katarzyna, Zhukov Igor, Maszota-Zieleniak Martyna, Michielin Olivier, Speiser Daniel E., Zoete Vincent, Derré Laurent, Rodziewicz-Motowidło Sylwia. Disulfide-Linked Peptides for Blocking BTLA/HVEM Binding. International Journal of Molecular Sciences 2020, 21(2): 636 (doi: 10.3390/ijms21020636)
-
Colizzi Vittorio, Mezzana Daniele, Ovseiko Pavel, V., Caiati Giovanni, Colonnello Claudia, Declich Andrea, Buchan Alastair M., Edmunds Laurel, Buzan Elena, Zerbini Luiz, Djilianov Dimitar, Schmidt Evanthia Kalpazidou, Bielawski Krzysztof P., Elster Doris, Solvato Maria, Alcantara Luiz C., Jr., Minutolo Antonella, Potesta Marina, Bachiddu Elena, Milano Maria J., Henderson Lorna R., Kiparoglou Vasiliki, Friesen Phoebe, Sheehan Mark, Moyankova Daniela, Rusanov Krasimir, Wium Martha, Raszczyk Izabela, Konieczny Igor, Gwizdala Jerzy P., Sledzik Karol, Barendziak Tanja, Birkholz Julia, Mueller Nicklas, Warrelmann Juergen, Meyer Ute, Filser Juliane, Barreto Fernando Khouri, Montesano Carla. Structural Transformation to Attain Responsible BIOSciences (STARBIOS2): Protocol for a Horizon 2020 Funded European Multicenter Project to Promote Responsible Research and Innovation. JMIR Research Protocols 2019, 8 (3): e11745 (doi: 10.2196/11745)
-
Dubiel Andrzej, Węgrzyn Katarzyna, Kupiński Adam P., Konieczny Igor. ClpAP protease is a universal factor that activates the parDE toxin-antitoxin system from a broad host range RK2 plasmid. Scientific Reports 2018, 8: 15287 ( doi: 10.1038/s41598-018-33726-y)
-
Bury Katarzyna, Węgrzyn Katarzyna, Konieczny Igor. Handcuffing reversal is facilitated by proteases and replication initiator monomers. Nucleic Acids Research 2017, 45(7): 3953-3966 (doi: 10.1093/nar/gkx166)
-
Karłowicz Anna, Węgrzyn Katarzyna, Gross Marta, Kaczyńska Dagmara, Ropelewska Małgorzata, Siemiątkowska Małgorzata, Bujnicki Janusz, Konieczny Igor. Defining the Crucial Domain and Amino Acid Residues in Bacterial Lon Protease for DNA Binding and Processing of DNA-interacting Substrates Journal of Biological Chemistry 2017, 292: 7507-7518 ( doi:10.1074/jbc.M116.766709)
-
Yano Hirokazu, Wegrzyn Katarzyna, Loftie-Eaton Wesley, Johnson Jenny Deckert Gail E., Rogers Linda M., Konieczny Igor, Top Eva M. Evolved plasmid-host interactions reduce plasmid interference cost. Molecular Microbiology 2016, 101(5): 743-756 (doi: 10.1111/mmi.13407)
- Wawrzycka Aleksandra, Gross Marta, Wasążnik Anna, Konieczny Igor. Plasmid replication initiator interactions with origin 13-mers and polymerase subunits contribute to strand-specific replisome assembly. Proceedings of the National Academy of Sciences of the United States of America 2015, 112(31): E4188-E4196 (doi: 10.1073/pnas.1504926112). *Award from PAN as the best Polish publication in microbiology in 2015
-
Węgrzyn Katarzyna, Fuentes-Perez Maria Eugenia, Bury Katarzyna, Rajewska Magdalena, Moreno-Herrero Fernando, Konieczny Igor. Sequence-specific interactions of Rep proteins with ssDNA in the AT-rich region of the plasmid replication origin. Nucleic Acids Research 2014, 42(12): 7807-7818 (doi: 10.1093/nar/gku453).
-
Kubik Sławomir, Węgrzyn Katarzyna, Pierechod Marcin, Konieczny Igor. Opposing effects of DNA on proteolysis of a replication initiator. Nucleic Acids Research 2012, 40(3): 1148-1159
-
Rajewska Magdalena, Węgrzyn Katarzyna, Konieczny Igor. AT-rich region and repeated sequences - the essential elements of replication origins of bacterial replicons. FEMS Microbiology Reviews 2012, 36(2): 408-434
-
Pierechod Marcin, Nowak Agnieszka, Saari Anna, Purta E., Bujnicki Janusz M., Konieczny I. Conformation of a plasmid replication initiator protein affects its proteolysis by ClpXP system. Protein Science 2009, 18(3): 637-649
- Rajewska M., Kowalczyk Ł., Konopa, G., Konieczny I. (2008) Specific mutations within the AT-rich region of a plasmid replication origin affect either origin opening or helicase loading. Proc Natl Acad Sci U S A 105(32): 11134-11139
-
Kowalczyk Ł., Rajewska M., Konieczny I. (2005) Positioning and the specific sequence of each 13-mer motif are critical for activity of the plasmid RK2 replication origin. Molecular Microbiol. 57(5): 1439-1449
-
Konieczny I. (2003) Strategies for helicase recruitment and loading in bacteria. EMBOReports 4(1):37-41
-
Jiang Y., Pacek M., Helinski D.R., Konieczny I., Toukdarian A. (2003) A multifunctional plasmid-encoded replication initiation protein both recruits and positions an active helicase at the replication origin. Proc Natl Acad Sci U S A 100(15): 8692-8697
-
Konieczny I., Liberek K. (2002) Cooperative action of Escherichia coli ClpB protein and DnaK chaperone in the activation of a replication initiation protein. J. Biol. Chem. 277(21): 18483-18488.
-
Jiang Y., Pogliano J., Helinski R.D., Konieczny I. (2002) ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Molecular Microbiol. 44(4): 971-979
-
Pacek M., Konopa G., Konieczny I. (2001) DnaA box sequences as the site for helicase delivery during plasmid RK2 replication initiation in Escherichia coli. J. Biol. Chem. 276(26): 23639-23644
-
Caspi R., Pacek M., Consiglieri G., Helinski D.R., Toukdarian A., Konieczny I. (2001) A broad host range replicon with different requirements for replication initiation in three bacterial species. EMBO Journal 20(12): 3262-3271.
-
Caspi R., Helinski D.R., Pacek M., Konieczny I. (2000) Interactions of DnaA proteins from distantly related bacteria with the replication origin of the broad host range plasmid RK2. J. Biol. Chem. 275(24): 454-18461
-
Doran K.S., Helinski D.R., Konieczny I. (1999) A critical DnaA box directs the cooperative binding of the Escherichia coli DnaA protein to the plasmid RK2 replication origin. J. Biol. Chem. 274(25): 17918-17923
- Konieczny I., Doran K.S., Helinski D.R., Blasina A. (1997) Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J. Biol. Chem. 272(32): 20173-20178.
-
Konieczny I., Helinski D.R. (1997) The replication initiation protein of the broad-host-range plasmid RK2 is activated by the ClpX chaperone. Proc Natl Acad Sci U S A 94(26): 14378-14382